What is Poliovirus
- nonenveloped virus with single-stranded RNA (positive-sense +ssRNA)
- composed of an RNA genome and a protein capsid
- 7500 nucleotides long
- viral particle is 30 nm in diameter with icosahedral symmetry
- widely regarded as the simplest significant virus
What’s the structure of Poliovirus
The poliovirus genome encodes a single polyprotein of 220 kD, which is cotranslationally processed by the virus-encoded 2Apro and 3Cpro proteases, and protease-containing precursors. The primary proteolytic cleavage events divide the genome into three precursors: P1, P2, and P3. P1 is subsequently processed to yield the four structural proteins, multiple copies of which assemble to form the ∼30-nm diameter icosahedral virus particle.
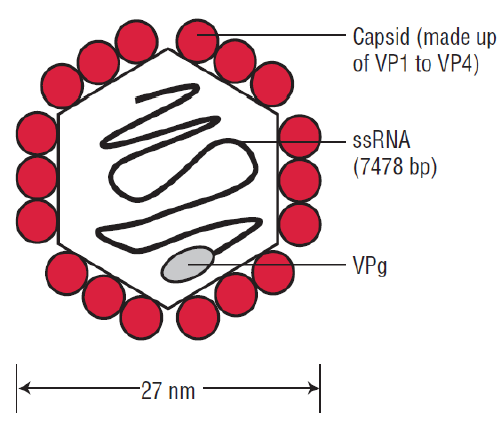
Figure 1.Structure of Polio Virus.
The P1 region is dispensable for replication, and can be replaced with a reporter gene to facilitate the direct quantification of genome replication by enzymatic or colorimetric assay.
The P2 and P3 regions contain the cis-acting nonstructural proteins required for virus replication, including the 2Apro and 3Cpro proteases, the 3Dpol RNA-dependent RNA polymerase, and VPg (3B), a 22 amino acid peptide that serves as a primer for the initiation of positive and negative strand genome replication and that is covalently attached to the virus genome.
The 5′ and 3′ noncoding regions (NCR) are extensively structured domains implicated in genome replication and translation, the latter via direct recruitment of ribosomes to the internal ribosome entry site (IRES) in the 5′ NCR.
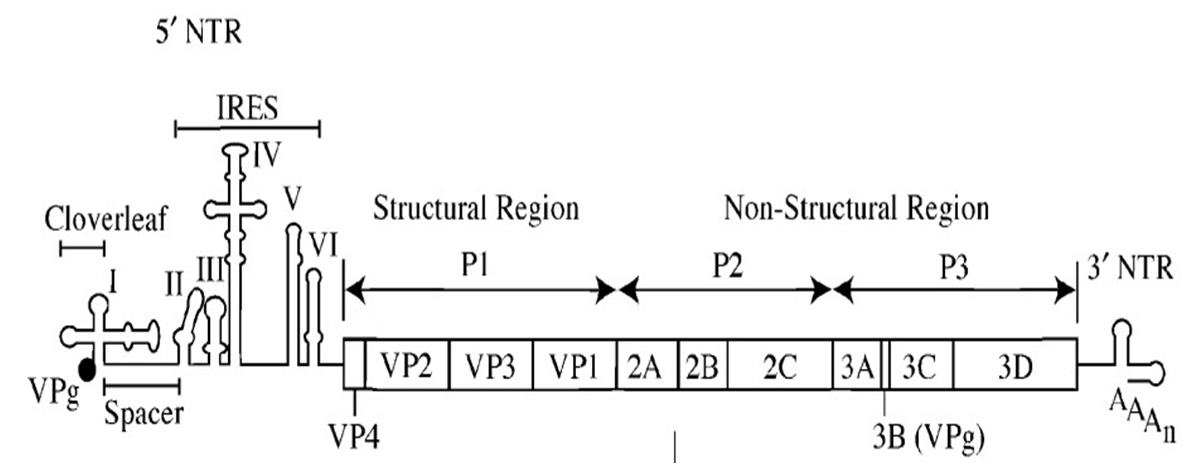
Figure 2.Poliovirus genome.png.
The IRES is preceded by an 88-nt ‘‘cloverleaf’’ (CL) structure that forms a ribonucleoprotein complex with a cellular protein (PCBP2) and the virus-encoded 3CDpro precursor.
the 3′ NCR is not absolutely required for replication—picornavirus 3′ NCR sequences can be functionally interchanged, and deletion of the 3′ NCR does not prevent virus replication. *cis*-acting replication elements (CRE) consist of short stem-loop structures predicted to contain an internal or terminal loop with three unpaired A nucleotides. To prime the initiation of replication, VPg must be uridylylated, a process involving the covalent addition of two uridine nucleotides to a tyrosine residue conserved in all picornaviruses.
Function of individual viral protein:
- 3Dpol, an RNA dependent RNA polymerase whose function is to make multiple copies of the viral RNA genome
- 2Apro and 3Cpro/3CDpro, proteases which cleave the viral polypeptide
- VPg (3B), a small protein that binds viral RNA and is necessary for synthesis of viral positive and negative strand RNA
- 2BC, 2B, 2C (an ATPase), 3AB, 3A, 3B proteins which comprise the protein complex needed for virus replication
- VP0, which is further cleaved into VP2 and VP4, VP1 and VP3, proteins of the viral capsid
How does Poliovirus replicate
The replication cycle of poliovirus is initiated by binding to the cell surface receptor CD155.
1.The virion forms a pore in the cell membrane through which viral RNA is released into the cytoplasm.
2.The 5' VPg can be cleaved off the genomic RNA by host TBP2, also called "unlinkase”. Translation of the viral RNA occurs by an IRES-mediated mechanism.
3.Poliovirus mRNA is translated as one long polypeptide. This polypeptide is then autocleaved by internal proteases into about 10 individual viral proteins.
4.The positive-sense RNA serves as template for complementary negative-strand synthesis, producing double-stranded replicative form (RF) RNA.
5.Many positive strand RNA copies are produced from the single negative strand. For synthesis of each negative-strand and positive-strand RNAs, VPg protein in the poliovirus works as a primer.
6.The newly synthesized positive-sense RNA molecules can serve as templates for translation of more viral proteins or can be enclosed in a capsid which ultimately generates progeny virions. Lysis of the infected cell results in release of infectious progeny virions.
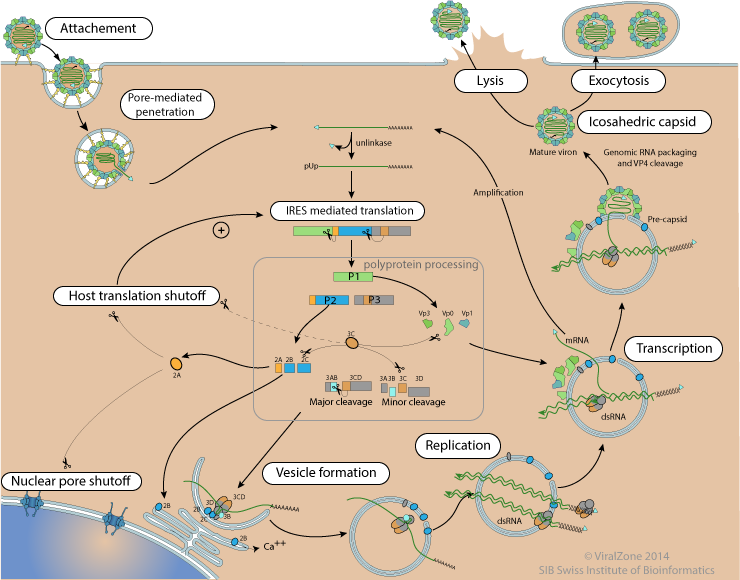
Figure 3.Poliovirus cycle.png.
Reference:
Echo Biosystems is committed to delivering high-quality proteins to support your scientific research. We have developed a series of high-quality virus proteins including glycoprotein, Matrix protein, Nucleoprotein, and Phosphoprotein to meet your research needs.
