I. Introduction
This protocol describes a method for coupling ligands containing hydrazine, hydrazide, aminooxy, or amine groups to aldehyde-containing particles. Aldehyde particles are reactive with hydrazine, hydrazide, or aminooxy derivatives, forming hydrazone or oxime linkages. Reactions with amine-containing molecules, such as proteins, can be carried out through a reductive amination process using sodium cyanoborohydride.
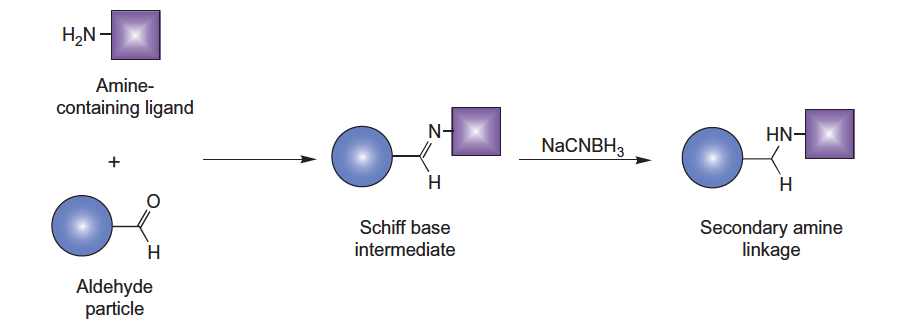
Figure. Aldehyde particles can be reacted with amine-containing proteins or other molecules.
II. Materials
- Aldehyde particles
- Coupling buffer (e.g., 10-mM sodium phosphate, pH 7.4; carbonate buffer at pH 10 for more efficient Schiff base formation with amine-containing molecules)
- Ligand to be coupled, containing hydrazine, hydrazide, aminooxy, or amine groups
- Sodium cyanoborohydride (NaCNBH3) for reductive amination with amine-containing ligands
- Blocking agent (e.g., glyceraldehyde, ethanolamine)
- Storage buffer with a preservative
III. Protocol
1. Wash aldehyde particles three times with coupling buffer. Buffers of higher pH value (e.g., carbonate buffer at pH 10) will result in more efficient Schiff base formation with amine-containing molecules than neutral pH conditions.
2. Mix the ligand to be coupled with the particle suspension.
3. For amine-containing ligands, carry out reductive amination using sodium cyanoborohydride.
- Add to the gel slurry 63 mg of solid sodium cyanoborohydride (NaCNBH3; MW = 62.84) or 0.2 ml of 5-M NaCNBH3 in 1-N NaOH with stirring.
- If the alkaline solution of cyanoborohydride is used, check the pH and readjust to 7.2, if necessary.
- Continue the reaction for 4 h to overnight at room temperature
4. React with mixing for 30 min at room temperature.
5. Wash the particles at least three times with water using centrifugation to remove excess reactants.
6. Block excess aldehyde groups, if necessary.
- Excess aldehyde groups on the support may be blocked by the addition of 50-mM hydroxylamine, which will react with the aldehydes to create stable oxime linkages.
- Unreacted amine residues remaining on the support can be blocked by the addition of a small aldehyde-containing compound, such as glyceraldehyde.
- Avoid the blocking of excess amine groups if a protein ligand or a ligand containing more than one amine has been immobilized, as the glyceraldehyde also will modify amines on the ligand.
- Wash the support once with an equal volume of 0.1-M glyceraldehyde dissolved in coupling buffer at pH 7.2, and then transfer the wet gel cake to a clean flask or vessel used for mixing the reaction. Add with stirring 10 ml of the 0.1-M glyceraldehyde solution along with 63 mg (or 0.2 ml of the 5-M NaCNBH3 solution) of sodium cyanoborohydride. Readjust the pH if necessary and continue the blocking reaction for 30 min at room temperature.
7. Wash and store the particles in a suitable buffer at neutral or slightly acidic pH containing a preservative. Thoroughly wash the particles with storage buffer (e.g., PBS, pH 7.5, or other suitable buffer) and resuspend them at 10 mg/ml in storage buffer containing a preservative.
IV. Considerations and Optimization
- pH: Use buffers of higher pH value e.g., carbonate buffer at pH 10) to result in more efficient Schiff base formation with amine-containing molecules than neutral pH conditions.
- Reductive Amination: For reactions with amine-containing molecules, use sodium cyanoborohydride for reductive amination.
- Blocking: Use appropriate blocking agents to block excess reactive groups.
- Washing: Ensure thorough washing to remove unreacted reagents and prevent nonspecific binding.
- Reaction Time and Temperature: A shorter coupling time may be sufficient for many ligands, but the optimal conditions to give acceptable yields for a particular ligand should be determined by doing a series of coupling reactions at different time points. Reactions at 4°C may also be done for thermally sensitive proteins or ligands, but reaction times likely will have to be extended to get the same level of coupling as a room temperature reaction.
V. Notes
- The optimal conditions for each step may need to be determined experimentally for each specific ligand and particle type.
- The degree of protein loading can be determined using a protein assay technique.
- For more information, consult the immobilization technology literature.
Interested in learning more about our particles with versatile functional groups for your conjugation? Visit Our Functional Polystyrene Particles to explore more.
Reference: Bioconjugate Techniques, 3rd Edition - July 25, 2013, Greg T. Hermanson
